This work is supported by Continuum Analytics and the XDATA Program as part of the Blaze Project
Very high performance isn’t about doing one thing well, it’s about doing nothing poorly.
This week I optimized the inter-node communication protocol used by
dask.distributed. It was a fun exercise in optimization that involved
several different and unexpected components. I separately had to deal with
Pickle, NumPy, Tornado, MsgPack, and compression libraries.
This blogpost is not advertising any particular functionality, rather it’s a story of the problems I ran into when designing and optimizing a protocol to quickly send both very small and very large numeric data between machines on the Python stack.
We care very strongly about both the many small messages case (thousands of 100 byte messages per second) and the very large messages case (100-1000 MB). This spans an interesting range of performance space. We end up with a protocol that costs around 5 microseconds in the small case and operates at 1-1.5 GB/s in the large case.
Identify a Problem
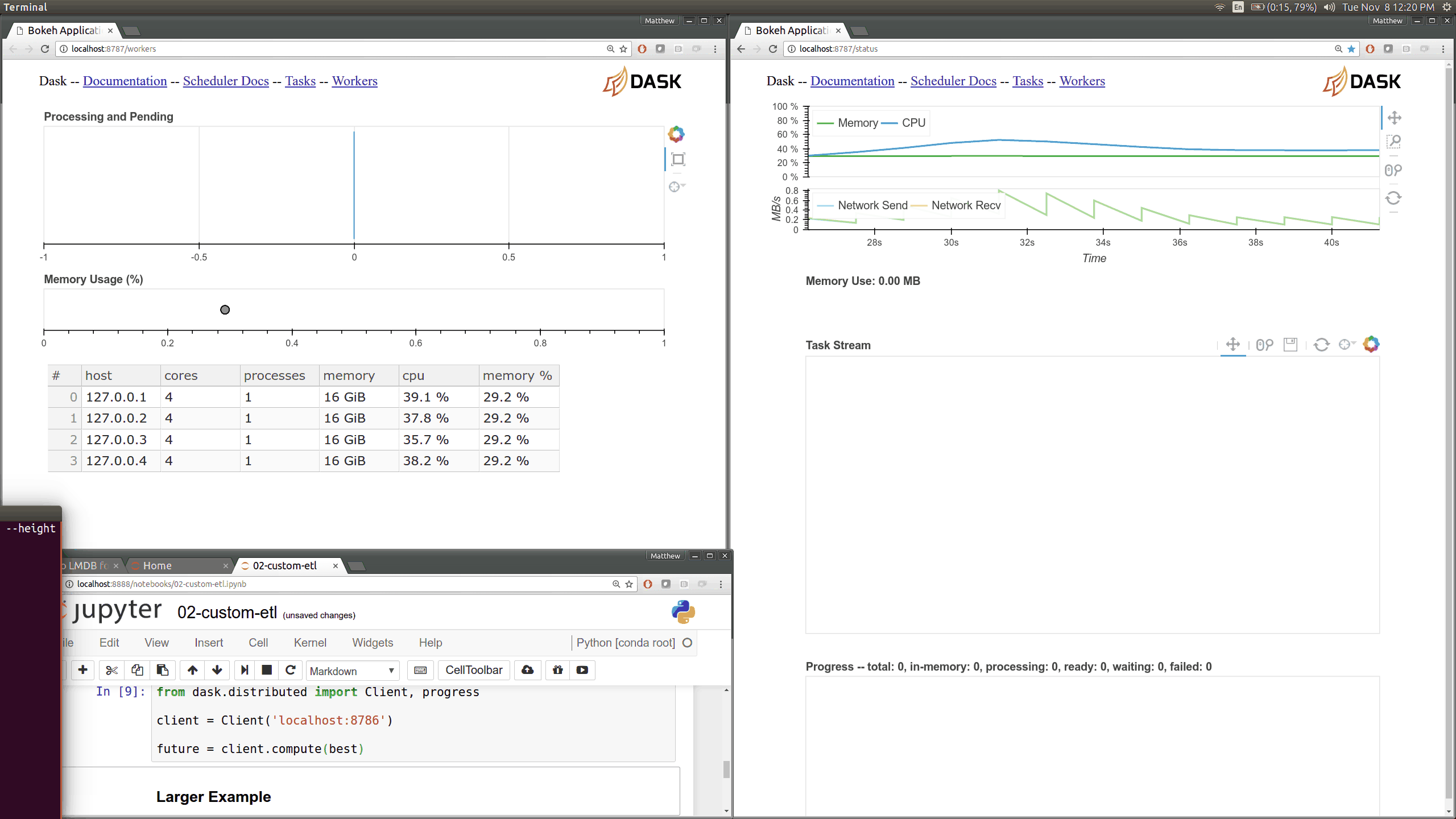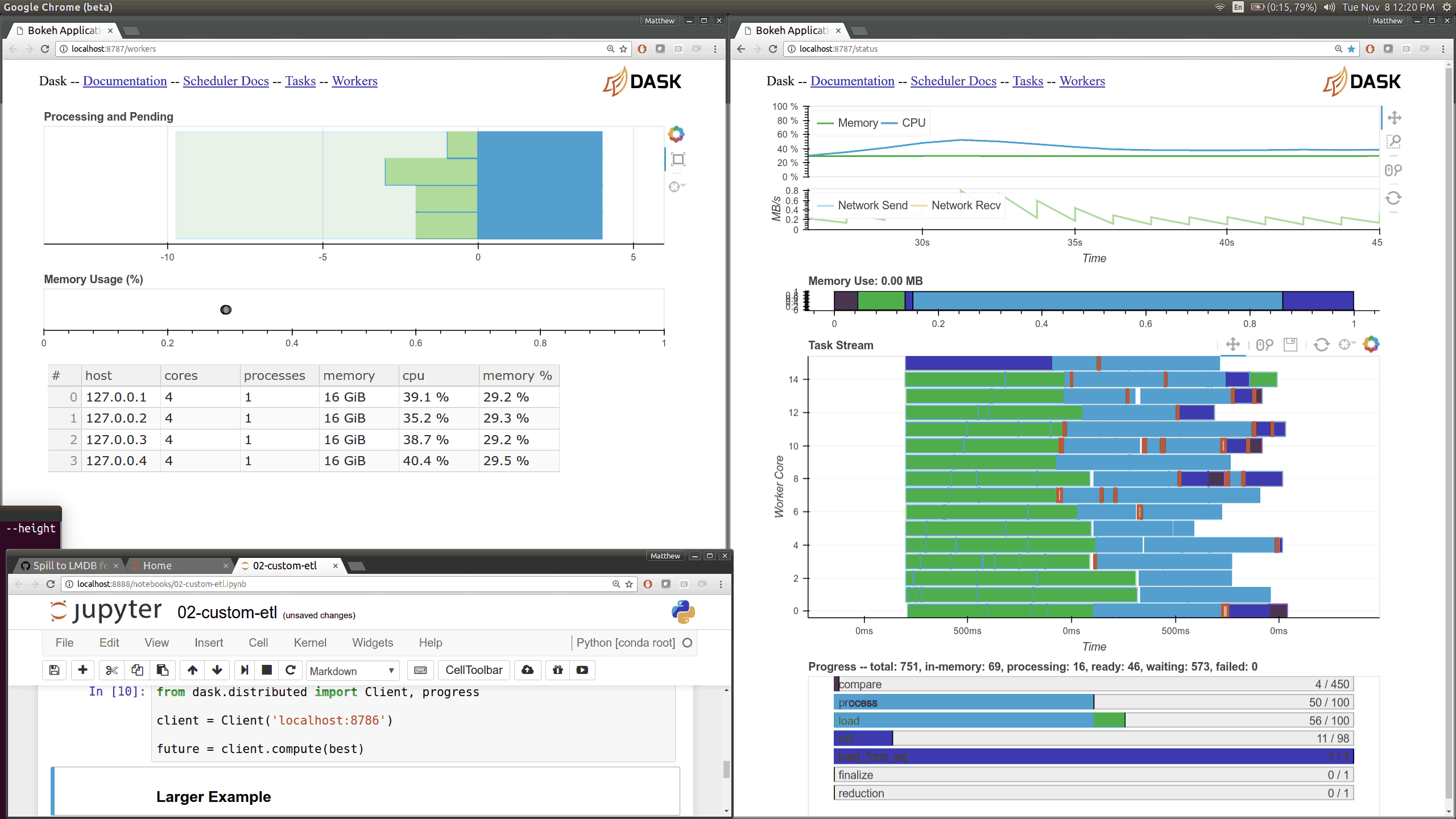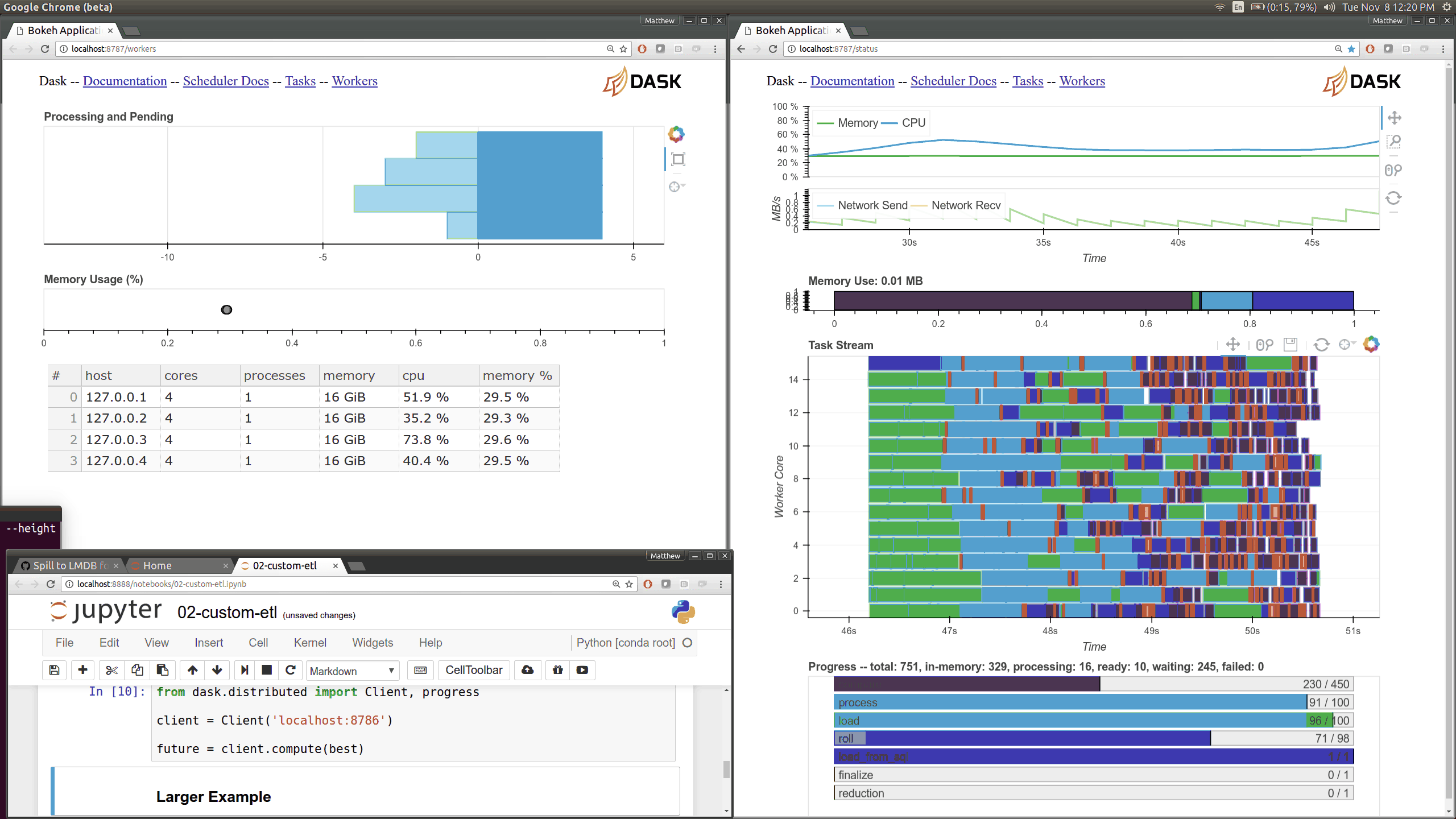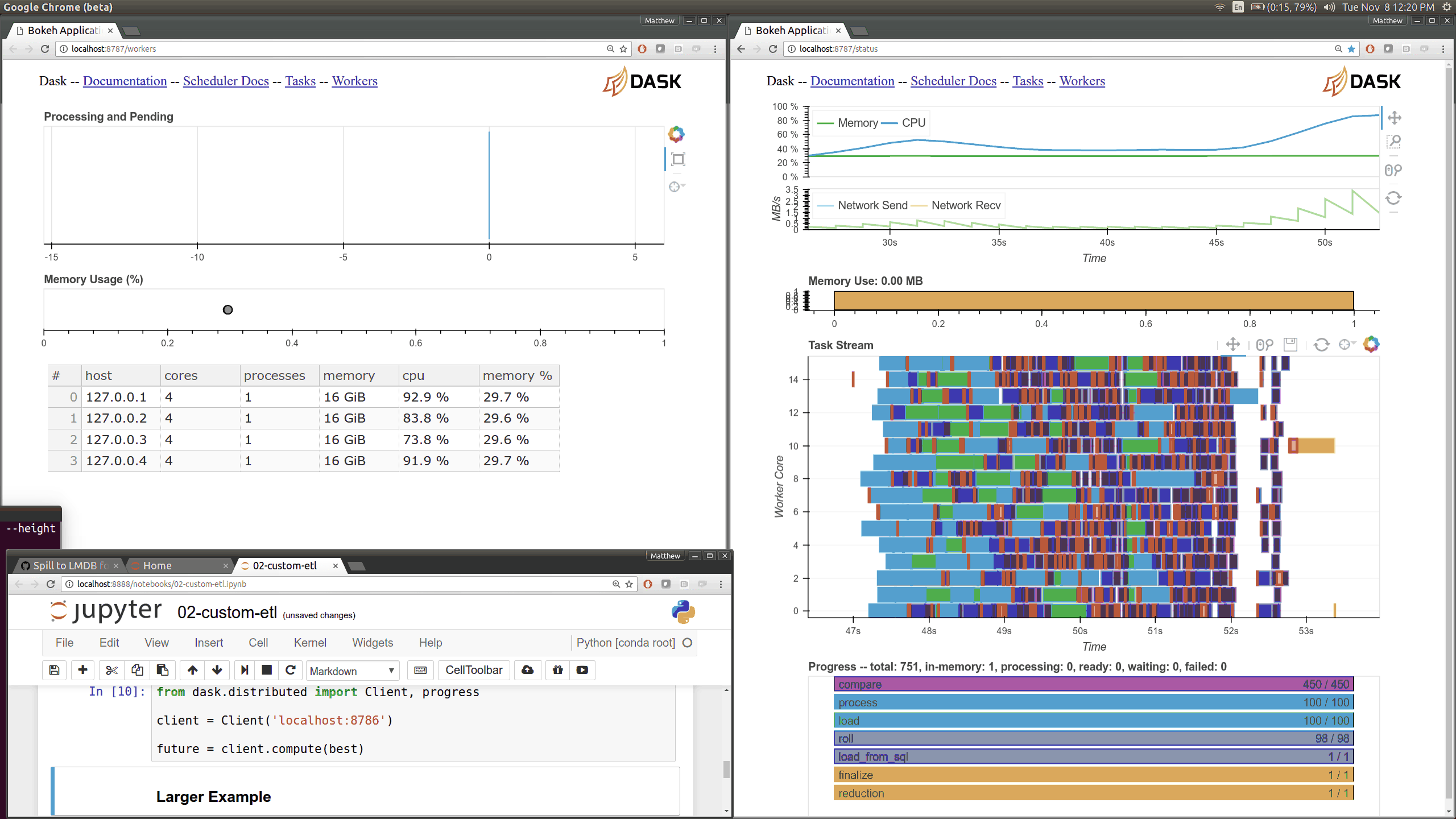
This came about as I was preparing a demo using dask.array on a distributed
cluster for a Continuum webinar. I noticed that my computations were taking
much longer than expected. The
Web UI quickly pointed
me to the fact that my machines were spending 10-20 seconds moving 30 MB chunks
of numpy array data between them. This is very strange because I was on
100MB/s network, and so I expected these transfers to happen in more like 0.3s
than 15s.
The Web UI made this glaringly apparent, so my first lesson was how valuable visual profiling tools can be when they make performance issues glaringly obvious. Thanks here goes to the Bokeh developers who helped the development of the Dask real-time Web UI.
Problem 1: Tornado’s sentinels
Dask’s networking is built off of Tornado’s TCP IOStreams.
There are two common ways to delineate messages on a socket, sentinel values
that signal the end of a message, and prefixing a length before every message.
Early on we tried both in Dask but found that prefixing a length before every
message was slow. It turns out that this was because TCP sockets try to batch
small messages to increase bandwidth. Turning this optimization off ended up
being an effective and easy solution, see the TCP_NODELAY parameter.
However, before we figured that out we used sentinels for a long time. Unfortunately Tornado does not handle sentinels well for large messages. At the receipt of every new message it reads through all buffered data to see if it can find the sentinel. This makes lots and lots of copies and reads through lots and lots of bytes. This isn’t a problem if your messages are a few kilobytes, as is common in web development, but it’s terrible if your messages are millions or billions of bytes long.
Switching back to prefixing messages with lengths and turning off the no-delay optimization moved our bandwidth up from 3MB/s to 20MB/s per node. Thanks goes to Ben Darnell (main Tornado developer) for helping us to track this down.
Problem 2: Memory Copies
A nice machine can copy memory at 5 GB/s. If your network is only 100 MB/s then you can easily suffer several memory copies in your system without caring. This leads to code that looks like the following:
socket.send(header + payload)
This code concatenates two bytestrings, header and payload before
sending the result down a socket. If we cared deeply about avoiding memory
copies then we might instead send these two separately:
socket.send(header)
socket.send(payload)
But who cares, right? At 5 GB/s copying memory is cheap!
Unfortunately this breaks down under either of the following conditions
- You are sloppy enough to do this multiple times
- You find yourself on a machine with surprisingly low memory bandwidth, like 10 times slower, as is the case on some EC2 machines.
Both of these were true for me but fortunately it’s usually straightforward to reduce the number of copies down to a small number (we got down to three), with moderate effort.
Problem 3: Unwanted Compression
Dask compresses all large messages with LZ4 or Snappy if they’re available. Unfortunately, if your data isn’t very compressible then this is mostly lost time. Doubly unforutnate is that you also have to decompress the data on the recipient side. Decompressing not-very-compressible data was surprisingly slow.
Now we compress with the following policy:
- If the message is less than 10kB, don’t bother
- Pick out five 10kB samples of the data and compress those. If the result isn’t well compressed then don’t bother compressing the full payload.
- Compress the full payload, if it doesn’t compress well then just send along the original to spare the receiver’s side from compressing.
In this case we use cheap checks to guard against unwanted compression. We also avoid any cost at all for small messages, which we care about deeply.
Problem 4: Cloudpickle is not as fast as Pickle
This was surprising, because cloudpickle mostly defers to Pickle for the easy stuff, like NumPy arrays.
In[1]:importnumpyasnpIn[2]:data=np.random.randint(0,255,dtype='u1',size=10000000)In[3]:importpickle,cloudpickleIn[4]:%timelen(pickle.dumps(data,protocol=-1))CPUtimes:user8.65ms,sys:8.42ms,total:17.1msWalltime:16.9msOut[4]:10000161In[5]:%timelen(cloudpickle.dumps(data,protocol=-1))CPUtimes:user20.6ms,sys:24.5ms,total:45.1msWalltime:44.4msOut[5]:10000161But it turns out that cloudpickle is using the Python implementation, while
pickle itself (or cPickle in Python 2) is using the compiled C implemenation.
Fortunately this is easy to correct, and a quick typecheck on common large
dataformats in Python (NumPy and Pandas) gets us this speed boost.
Problem 5: Pickle is still slower than you’d expect
Pickle runs at about half the speed of memcopy, which is what you’d expect from a protocol that is mostly just “serialize the dtype, strides, then tack on the data bytes”. There must be an extraneous memory copy in there.
See issue 7544
Problem 6: MsgPack is bad at large bytestrings
Dask serializes most messages with MsgPack, which is ordinarily very fast. Unfortunately the MsgPack spec doesn’t support bytestrings greater than 4GB (which do come up for us) and the Python implementations don’t pass through large bytestrings very efficiently. So we had to handle large bytestrings separately. Any message that contains bytestrings over 1MB in size will have them stripped out and sent along in a separate frame. This both avoids the MsgPack overhead and avoids a memory copy (we can send the bytes directly to the socket).
Problem 7: Tornado makes a copy
Sockets on Windows don’t accept payloads greater than 128kB in size. As a result Tornado chops up large messages into many small ones. On linux this memory copy is extraneous. It can be removed with a bit of logic within Tornado. I might do this in the moderate future.
Results
We serialize small messages in about 5 microseconds (thanks msgpack!) and move large bytes around in the cost of three memory copies (about 1-1.5 GB/s) which is generally faster than most networks in use.
Here is a profile of sending and receiving a gigabyte-sized NumPy array of random values through to the same process over localhost (500 MB/s on my machine.)
381360 function calls (381323 primitive calls) in 1.451 seconds
Ordered by: internal time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.366 0.366 0.366 0.366 {built-in method dumps}
8 0.289 0.036 0.291 0.036 iostream.py:360(write)
15353 0.228 0.000 0.228 0.000 {method 'join' of 'bytes' objects}
15355 0.166 0.000 0.166 0.000 {method 'recv' of '_socket.socket' objects}
15362 0.156 0.000 0.398 0.000 iostream.py:1510(_merge_prefix)
7759 0.101 0.000 0.101 0.000 {method 'send' of '_socket.socket' objects}
17/14 0.026 0.002 0.686 0.049 gen.py:990(run)
15355 0.021 0.000 0.198 0.000 iostream.py:721(_read_to_buffer)
8 0.018 0.002 0.203 0.025 iostream.py:876(_consume)
91 0.017 0.000 0.335 0.004 iostream.py:827(_handle_write)
89 0.015 0.000 0.217 0.002 iostream.py:585(_read_to_buffer_loop)
122567 0.009 0.000 0.009 0.000 {built-in method len}
15355 0.008 0.000 0.173 0.000 iostream.py:1010(read_from_fd)
38369 0.004 0.000 0.004 0.000 {method 'append' of 'list' objects}
7759 0.004 0.000 0.104 0.000 iostream.py:1023(write_to_fd)
1 0.003 0.003 1.451 1.451 ioloop.py:746(start)
Dominant unwanted costs include the following:
- 400ms: Pickling the NumPy array
- 400ms: Bytestring handling within Tornado
After this we’re just bound by pushing bytes down a wire.
Conclusion
Writing fast code isn’t about writing any one thing particularly well, it’s about mitigating everything that can get in your way. As you approch peak performance, previously minor flaws suddenly become your dominant bottleneck. Success here depends on frequent profiling and keeping your mind open to unexpected and surprising costs.